Technology focus: MIBI-TOF#
This notebook will present a rough overview of the plotting functionalities that spatialdata implements for Visium data.
Loading the data#
Please download the data from here: MIBI-TOF dataset and adjust the variable containing the location of the .zarr file.
mibitof_zarr_path = "./mibitof.zarr"
import spatialdata as sd
mibitof_sdata = sd.read_zarr(mibitof_zarr_path)
mibitof_sdata
SpatialData object with:
├── Images
│ ├── 'point8_image': SpatialImage[cyx] (3, 1024, 1024)
│ ├── 'point16_image': SpatialImage[cyx] (3, 1024, 1024)
│ └── 'point23_image': SpatialImage[cyx] (3, 1024, 1024)
├── Labels
│ ├── 'point8_labels': SpatialImage[yx] (1024, 1024)
│ ├── 'point16_labels': SpatialImage[yx] (1024, 1024)
│ └── 'point23_labels': SpatialImage[yx] (1024, 1024)
└── Table
└── AnnData object with n_obs × n_vars = 3309 × 36
obs: 'row_num', 'point', 'cell_id', 'X1', 'center_rowcoord', 'center_colcoord', 'cell_size', 'category', 'donor', 'Cluster', 'batch', 'library_id'
uns: 'spatialdata_attrs'
obsm: 'X_scanorama', 'X_umap', 'spatial': AnnData (3309, 36)
with coordinate systems:
▸ 'point8', with elements:
point8_image (Images), point8_labels (Labels)
▸ 'point16', with elements:
point16_image (Images), point16_labels (Labels)
▸ 'point23', with elements:
point23_image (Images), point23_labels (Labels)
Visualise the data#
We’re going to create a naiive visualisation of the data, overlaying the segmented cells and the tissue images. For this, we need to load the spatialdata_plot library which extends the sd.SpatialData object with the .pl module.
import spatialdata_plot
mibitof_sdata.pl.render_images().pl.render_labels().pl.show()
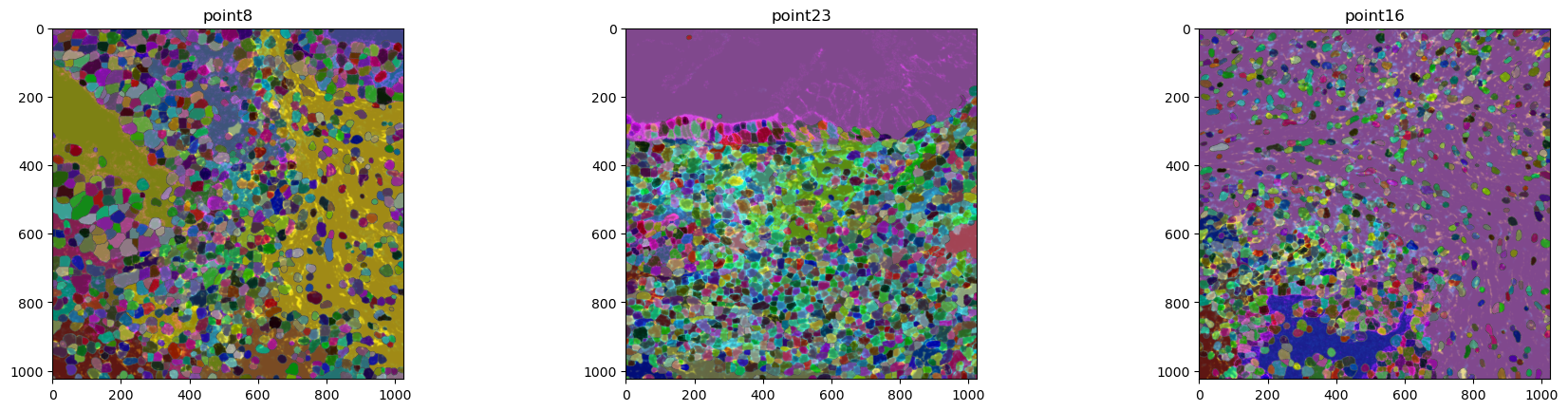
We can see that the data contains three coordinate systems (point8, point16 and point23) with image and cell segmentation information each. When giving no further parameters, one panel is generated per coordinate system with the members that have been specified in the function call. While it is hard to see, the cell labels overlay the tissue image nicely. To better show this, we will plot the data on individual ax objects.
import matplotlib.pyplot as plt
fig, axs = plt.subplots(ncols=3, nrows=1, figsize=(10, 4))
mibitof_sdata_subset = mibitof_sdata.pp.get_elements("point8")
mibitof_sdata_subset.pl.render_images().pl.show(ax=axs[0], title="image")
mibitof_sdata_subset.pl.render_labels().pl.show(ax=axs[1], title="labels")
mibitof_sdata_subset.pl.render_images().pl.render_labels().pl.show(ax=axs[2], title="both")
plt.tight_layout()
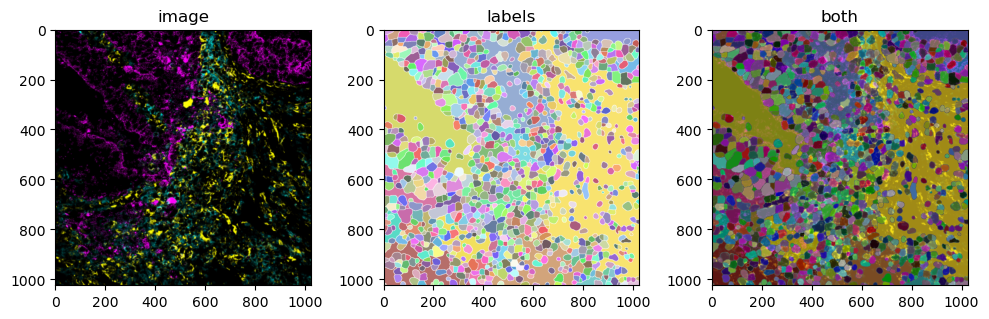
However, the segmentation masks are all shown in random colors since we have not provided any information on what they should encode. Such information can be found in the Table attribute (which is an anndata.AnnData table) of the SpatialData object,either in the data itself or the obs attribute.
mibitof_sdata["table"].to_df().head(3)
| ASCT2 | ATP5A | CD11c | CD14 | CD3 | CD31 | CD36 | CD39 | CD4 | CD45 | ... | NRF2p | NaKATPase | PD1 | PKM2 | S6p | SDHA | SMA | VDAC1 | XBP1 | vimentin | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 9376-1 | -0.099024 | 0.047260 | -0.076592 | -0.160894 | -0.094239 | -0.059072 | -0.048790 | -0.080359 | -0.181162 | -0.177686 | ... | -0.063972 | -0.062695 | -0.078785 | -0.189159 | -0.233837 | 0.038624 | -0.502865 | -0.016736 | 0.055549 | -0.410221 |
| 9377-1 | -0.081413 | 0.025841 | -0.062929 | 0.071132 | -0.086687 | -0.072516 | 0.209030 | 0.067253 | 0.088813 | -0.164108 | ... | 0.116562 | 0.115541 | -0.075863 | 0.024126 | -0.318994 | -0.043148 | -0.120517 | -0.058352 | -0.197411 | -0.179946 |
| 9378-1 | -0.100959 | -0.203419 | -0.055992 | -0.134076 | -0.066981 | -0.047282 | -0.044181 | -0.170404 | -0.045016 | -0.110186 | ... | -0.031722 | -0.109969 | -0.060554 | -0.244716 | -0.218434 | -0.172760 | -0.301259 | -0.210778 | 0.042039 | -0.266521 |
3 rows × 36 columns
mibitof_sdata["table"].obs.head(3)
| row_num | point | cell_id | X1 | center_rowcoord | center_colcoord | cell_size | category | donor | Cluster | batch | library_id | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 9376-1 | 9479 | 8 | 2 | 65222.0 | 37.0 | 6.0 | 474.0 | carcinoma | 90de | Epithelial | 1 | point8_labels |
| 9377-1 | 9480 | 8 | 4 | 65224.0 | 314.0 | 3.0 | 126.0 | carcinoma | 90de | Epithelial | 1 | point8_labels |
| 9378-1 | 9481 | 8 | 5 | 65225.0 | 407.0 | 6.0 | 398.0 | carcinoma | 90de | Epithelial | 1 | point8_labels |
Color the segmentation masks by a categorical variable#
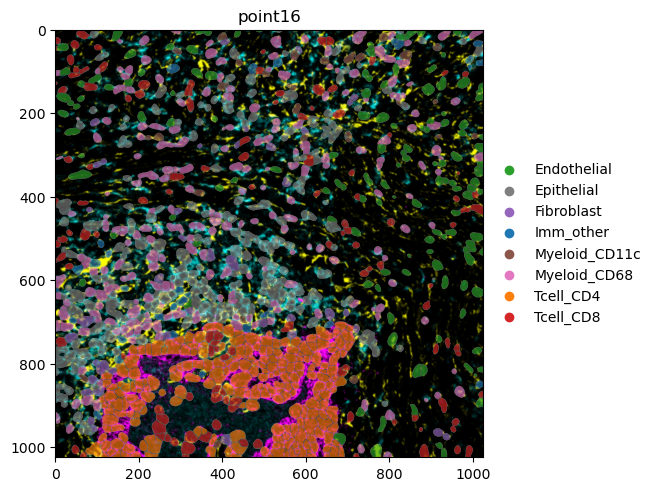
To use this information in our plot, we provide the column-name to be used for the color-enocoding to color in render_labels(). Here, spatialdata-plot automatically differentiates between categorical and numerical columns. Furthermore, we subset the data to only one coordinate system.
mibitof_sdata.pp.get_elements("point16").pl.render_images().pl.render_labels(color="Cluster").pl.show()
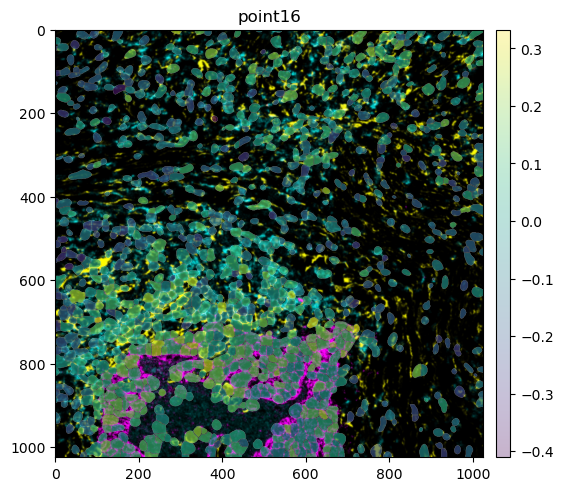
mibitof_sdata.pp.get_elements("point16").pl.render_images().pl.render_labels(color="ASCT2").pl.show()