Integrate/aggregate signals across spatial layers#
In this notebook, we will describe some usage principles for the aggregate method.
Let’s first import some useful libraries and create some dummy data to show the example.
%load_ext autoreload
%autoreload 2
%load_ext jupyter_black
import os
os.environ["USE_PYGEOS"] = "0"
import spatialdata_plot
from spatialdata.datasets import blobs
sdata = blobs()
sdata
SpatialData object with:
├── Images
│ ├── 'blobs_image': SpatialImage[cyx] (3, 512, 512)
│ └── 'blobs_multiscale_image': MultiscaleSpatialImage[cyx] (3, 512, 512), (3, 256, 256), (3, 128, 128)
├── Labels
│ ├── 'blobs_labels': SpatialImage[yx] (512, 512)
│ └── 'blobs_multiscale_labels': MultiscaleSpatialImage[yx] (512, 512), (256, 256), (128, 128)
├── Points
│ └── 'blobs_points': DataFrame with shape: (<Delayed>, 4) (2D points)
├── Shapes
│ ├── 'blobs_circles': GeoDataFrame shape: (5, 2) (2D shapes)
│ ├── 'blobs_multipolygons': GeoDataFrame shape: (2, 1) (2D shapes)
│ └── 'blobs_polygons': GeoDataFrame shape: (5, 1) (2D shapes)
└── Tables
└── 'table': AnnData (26, 3)
with coordinate systems:
▸ 'global', with elements:
blobs_image (Images), blobs_multiscale_image (Images), blobs_labels (Labels), blobs_multiscale_labels (Labels), blobs_points (Points), blobs_circles (Shapes), blobs_multipolygons (Shapes), blobs_polygons (Shapes)
import matplotlib.pyplot as plt
fig, axs = plt.subplots(ncols=3, figsize=(12, 3))
sdata.pl.render_images("blobs_image").pl.show(ax=axs[0])
sdata.pl.render_labels("blobs_labels").pl.show(ax=axs[1])
sdata.pl.render_points("blobs_points").pl.show(ax=axs[2])
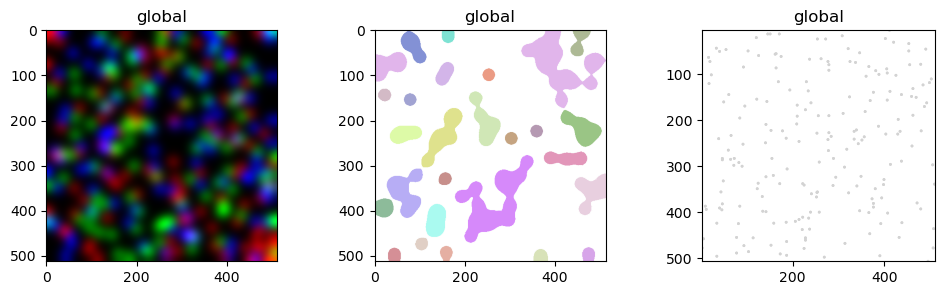
We can do various type of aggregations:
aggregate images by labels
aggregate points by shapes
aggregate shapes by shapes
Aggregations between mixed raster and non-raster types are currently not supported (but will be).
The API function for aggregation is spatialdata.aggregate. It is also possible to perform aggregation using the convenience method spatialdata.SpatialData.aggregate, which simply calls the previous one and automatically fills some values (values_sdata and by_sdata) with self; we will see this below.
Let’s start with aggregation of images by labels. This can be achieved with one line of code:
Aggregating images by labels#
sdata_im = sdata.aggregate(values="blobs_image", by="blobs_labels", agg_func="mean")
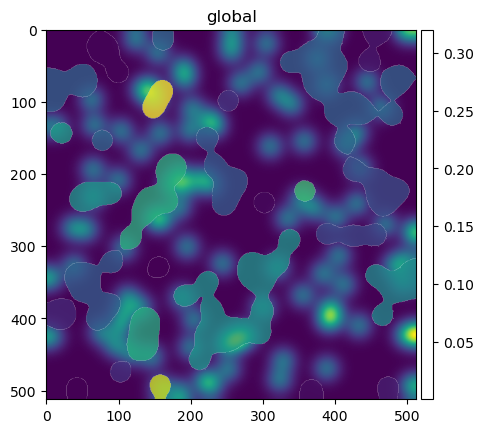
As default, the aggregation function is sum but it can be changed with the agg_func parameter. We can inspect the table inside the returned SpatialData object to confirm that the operation results in retrieving the mean intensity of each image channel within the boundaries of the labels.
We can also visualize the results with spatialdata-plot. By overlaying the labels with the channel where we performed aggregation, we can better appreciate the results. Labels that overlap with high intensity of the channels have indeed a higher mean intensity.
The features of the new table are the following:
sdata_im["table"].var_names
Index(['channel_0_mean', 'channel_1_mean', 'channel_2_mean'], dtype='object')
ax = plt.gca()
sdata.pl.render_images("blobs_image", cmap="viridis", channel=1).pl.show(ax=ax)
sdata_im.pl.render_labels(color="channel_1_mean", fill_alpha=0.5).pl.show(ax=ax)
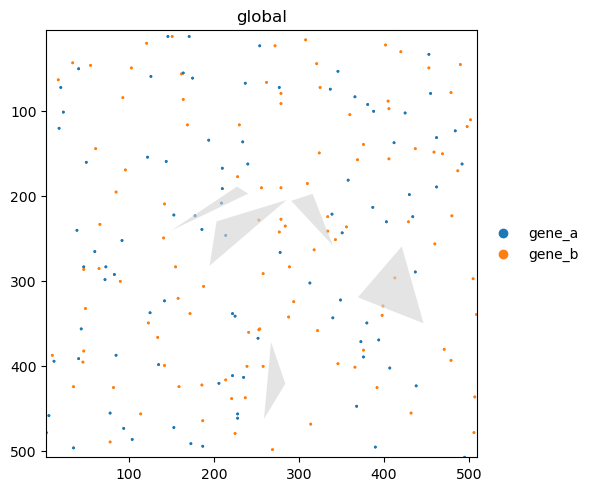
We can also aggregate points by shapes. For example, let’s count the number of points that overlap each shape.
Here we see the points and shapes that we will use for aggregation.
Aggregating points by shapes#
sdata["blobs_points"].compute()
| x | y | genes | instance_id | |
|---|---|---|---|---|
| 0 | 46 | 395 | gene_b | 9 |
| 1 | 334 | 224 | gene_b | 7 |
| 2 | 221 | 438 | gene_b | 3 |
| 3 | 44 | 356 | gene_a | 9 |
| 4 | 103 | 49 | gene_b | 4 |
| ... | ... | ... | ... | ... |
| 195 | 381 | 92 | gene_a | 8 |
| 196 | 188 | 306 | gene_b | 5 |
| 197 | 368 | 447 | gene_a | 7 |
| 198 | 23 | 101 | gene_a | 6 |
| 199 | 144 | 159 | gene_a | 6 |
200 rows × 4 columns
sdata.pl.render_points(color="genes").pl.render_shapes("blobs_polygons", fill_alpha=0.6).pl.show()
The value_key parameters specifies which columns of the points dataframe will be aggregated.
sdata_shapes = sdata.aggregate(values="blobs_points", by="blobs_polygons", value_key="genes", agg_func="count")
sdata_shapes
SpatialData object with:
├── Shapes
│ └── 'blobs_polygons': GeoDataFrame shape: (5, 1) (2D shapes)
└── Tables
└── 'table': AnnData (5, 2)
with coordinate systems:
▸ 'global', with elements:
blobs_polygons (Shapes)
Let’s color by the var value gene_b of the aggregate table (that is, we color by the numbers of points of type gene_b inside each shape).
ax = plt.gca()
sdata.pl.render_points(color="genes").pl.show(ax=ax)
sdata_shapes.pl.render_shapes(color="gene_b", fill_alpha=0.7).pl.show(ax=ax)
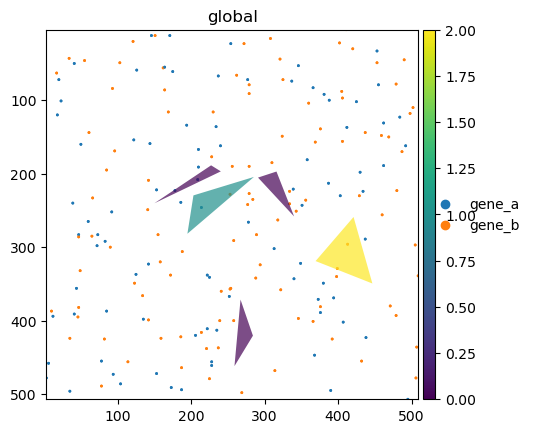
The colormap represents the number of counts for the selected variable (gene “b”). The rightmost polygon has correctly a value of 2, as there are 2 transcripts (points) of type “b” overlapping the polygon area.
Aggregating shapes by shapes, and information from different locations#
In this example let’s show four things:
aggregating shapes by shapes
aggregating layers from different
SpatialDataobjectsaggregating signals from different location within the same
SpatialDataobject (value_keyparameter).explain the difference of the
aggregate()function and theaggregate()method
To do so, let’s create two SpatialData objects:
one with the
blobs_circlesgeometries, but adding some extra annotations, both as newGeoDataFramecolumns, and in anAnnDatatable;one wiht two large rectangles that we will use to aggregate the circles by.
Creating the circles object#
sdata.pl.render_shapes("blobs_circles").pl.show()
import numpy as np
import pandas as pd
from anndata import AnnData
from numpy.random import default_rng
from spatialdata import SpatialData
from spatialdata.models import TableModel
RNG = default_rng(42)
adata_circles = AnnData(
RNG.normal(size=(5, 3)),
var=pd.DataFrame(index=["gene_h", "gene_k", "gene_l"]),
obs=pd.DataFrame(
{
"categorical": ["a", "a", "b", "c", "d"],
"region": "blobs_circles",
"instance_id": np.arange(5),
}
),
)
adata_circles.obs["categorical"] = adata_circles.obs["categorical"].astype("category")
adata_circles.obs["region"] = adata_circles.obs["region"].astype("category")
adata_circles = TableModel.parse(adata_circles, region="blobs_circles", region_key="region", instance_key="instance_id")
sdata_circles = SpatialData(shapes={"blobs_circles": sdata["blobs_circles"]}, tables={"table": adata_circles})
# let's add two numerical columns to the GeoDataFrame
sdata_circles["blobs_circles"]["feature_m"] = RNG.normal(size=(5))
sdata_circles["blobs_circles"]["feature_n"] = RNG.normal(size=(5))
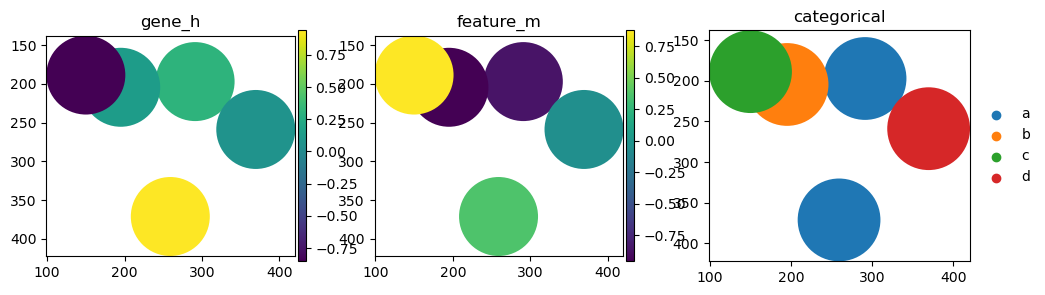
import matplotlib.pyplot as plt
fig, axs = plt.subplots(ncols=3, figsize=(12, 3))
sdata_circles.pl.render_shapes(color="gene_h").pl.show(ax=axs[0], title="gene_h")
sdata_circles.pl.render_shapes(color="feature_m").pl.show(ax=axs[1], title="feature_m")
sdata_circles.pl.render_shapes(color="categorical").pl.show(ax=axs[2], title="categorical")
Creating the squares object#
import geopandas as gpd
from shapely import linearrings, polygons
from spatialdata.models import ShapesModel
def _make_squares(centroid_coordinates: np.ndarray, half_widths: list[float]) -> polygons:
linear_rings = []
for centroid, half_width in zip(centroid_coordinates, half_widths):
min_coords = centroid - half_width
max_coords = centroid + half_width
linear_rings.append(
linearrings(
[
[min_coords[0], min_coords[1]],
[min_coords[0], max_coords[1]],
[max_coords[0], max_coords[1]],
[max_coords[0], min_coords[1]],
]
)
)
s = polygons(linear_rings)
polygon_series = gpd.GeoSeries(s)
cell_polygon_table = gpd.GeoDataFrame(geometry=polygon_series)
return ShapesModel.parse(cell_polygon_table)
sdata_squares = SpatialData(
shapes={"squares": _make_squares(np.atleast_2d([[100, 200], [400, 200]]), half_widths=[100, 80])}
)
ax = plt.gca()
sdata_squares.pl.render_shapes("squares", na_color="red", alpha=0.5).pl.show(ax=ax)
sdata_circles.pl.render_shapes("blobs_circles", alpha=0.5).pl.show(ax=ax)
We will now aggregate the various quantities. Notice how the value_key can be used to aggregate values that are located in different places in the SpatialData object:
matrix
Xof theAnnDatatable (names given by.var_names);.obsDataFrameof theAnnDatatable;columns of the
GeoDataFrame.
Notice also that the API deal both with numerical and categorical values, and can aggregate multiple numerical columns at the same time.
Case: value_key referring to var_names#
from spatialdata import aggregate
sdata_gene_exp = aggregate(
values_sdata=sdata_circles,
by_sdata=sdata_squares,
values="blobs_circles",
by="squares",
value_key=["gene_h", "gene_k"],
table_name="table",
)
print(sdata_gene_exp)
print()
print(sdata_gene_exp["table"].var_names)
SpatialData object with:
├── Shapes
│ └── 'squares': GeoDataFrame shape: (2, 1) (2D shapes)
└── Tables
└── 'table': AnnData (2, 2)
with coordinate systems:
▸ 'global', with elements:
squares (Shapes)
Index(['gene_h', 'gene_k'], dtype='object')
Case: value_key referring to GeoDataFrame columns#
sdata_feature = aggregate(
values_sdata=sdata_circles,
by_sdata=sdata_squares,
values="blobs_circles",
by="squares",
value_key="feature_m",
)
print(sdata_feature)
print()
print(sdata_feature["table"].var_names)
SpatialData object with:
├── Shapes
│ └── 'squares': GeoDataFrame shape: (2, 1) (2D shapes)
└── Tables
└── 'table': AnnData (2, 1)
with coordinate systems:
▸ 'global', with elements:
squares (Shapes)
Index(['feature_m'], dtype='object')
Case: value_key referring to obs columns#
sdata_categorical = aggregate(
values_sdata=sdata_circles,
by_sdata=sdata_squares,
values="blobs_circles",
by="squares",
value_key="categorical",
)
print(sdata_categorical)
print()
print(sdata_categorical["table"].var_names)
SpatialData object with:
├── Shapes
│ └── 'squares': GeoDataFrame shape: (2, 1) (2D shapes)
└── Tables
└── 'table': AnnData (2, 4)
with coordinate systems:
▸ 'global', with elements:
squares (Shapes)
Index(['a', 'b', 'c', 'd'], dtype='object')
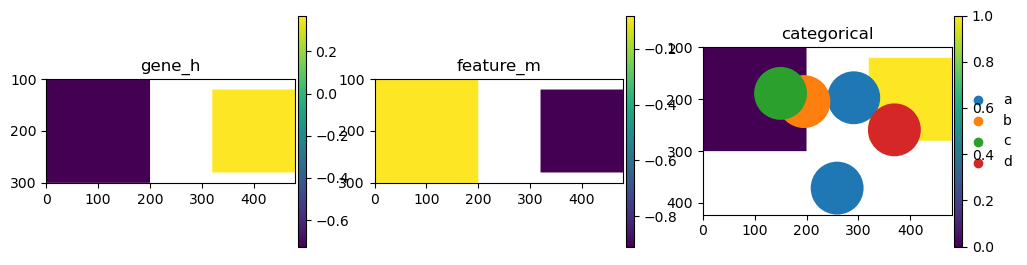
import matplotlib.pyplot as plt
fig, axs = plt.subplots(ncols=3, figsize=(12, 3))
sdata_gene_exp.pl.render_shapes("squares", color="gene_h", alpha=0.5).pl.show(ax=axs[0], title="gene_h")
sdata_feature.pl.render_shapes("squares", color="feature_m", alpha=0.5).pl.show(ax=axs[1], title="feature_m")
sdata_categorical.pl.render_shapes("squares", color="a", alpha=0.5).pl.show(ax=axs[2], title="categorical")
sdata_circles.pl.render_shapes(color="categorical").pl.show(ax=axs[2], title="categorical")
The method vs the function#
Above we used the aggregate() function. The method version is equivalent and it is a convenience function that simply fills in eventually missing values of values_sdata and by_sdata with self.
So these two functions are equivalent:
res0 = aggregate(
values_sdata=sdata_circles,
by_sdata=sdata_squares,
values="blobs_circles",
by="squares",
value_key="feature_m",
)
res1 = sdata_circles.aggregate(
by_sdata=sdata_squares,
values="blobs_circles",
by="squares",
value_key="feature_m",
)
One can also directly pass to values or by a SpatialElement. So also this is an equivalent call, except for the fact that now the string squares is never passed to aggregate(), so it will be used a default name for the name of the SpatialElement
res2 = sdata_circles.aggregate(
by=sdata_squares["squares"],
values="blobs_circles",
value_key="feature_m",
)
from anndata.tests.helpers import assert_equal
assert_equal(res0["table"], res1["table"])
assert_equal(res0["table"].X, res2["table"].X)
res0["table"].obs
| instance_id | region | |
|---|---|---|
| 0 | 0 | squares |
| 1 | 1 | squares |
res2["table"].obs
| instance_id | region | |
|---|---|---|
| 0 | 0 | by |
| 1 | 1 | by |
