Deep learning example on image tiles#
We will show, as an example, how to train a Dense Net which predicts cell types Xenium data from an associated H&E image.
In particular this example shows that:
We can easily access and combine images and annotations across different technologies. For the sake of the example here we use the H&E image from Visium data, and the cell type information from overlapping Xenium data. Remarkably, the two modalities are spatially aligned via an affine transformation.
We generate image tiles with full control of the spatial extent and the pixel resolution.
We interface with popular frameworks for deep learning: Monai and PyTorch Lightning.
%load_ext autoreload
%autoreload 2
%load_ext jupyter_black
import os
from typing import Dict
import numpy as np
import pandas as pd
import pytorch_lightning as pl
import scanpy as sc
import torch
import torch.multiprocessing as mp
import torch.nn.functional as F
import torchvision
from anndata import AnnData
from monai.networks.nets import DenseNet121
from pytorch_lightning import LightningDataModule
from pytorch_lightning.callbacks import LearningRateMonitor
from pytorch_lightning.callbacks.progress import TQDMProgressBar
from pytorch_lightning.loggers import CSVLogger
from spatial_image import SpatialImage
from spatialdata import SpatialData, read_zarr, transform
from spatialdata.dataloader.datasets import ImageTilesDataset
from spatialdata.transformations import get_transformation
from torch.nn import CrossEntropyLoss
from torch.optim import Adam
from torch.utils.data import DataLoader
from tqdm import tqdm
mp.set_start_method("spawn", force=True)
from napari_spatialdata import Interactive
Preparing the data#
Getting the Zarr files#
You can download the processed Visium and Xenium data (already aligned and Xenium with already the celltype information) from here: Visium dataset, Xenium dataset. In alternative you can obtain the data by running this analysis notebook.
Please rename the files to visium_aligned.zarr and xenium_aligned.zarr and place them in the same folder as this notebook (or use symlinks to make the data accessible).
XENIUM_SDATA_PATH = "xenium_aligned.zarr"
VISIUM_SDATA_PATH = "visium_aligned.zarr"
assert os.path.isdir(XENIUM_SDATA_PATH)
assert os.path.isdir(VISIUM_SDATA_PATH)
xenium_sdata = read_zarr(XENIUM_SDATA_PATH)
visium_sdata = read_zarr(VISIUM_SDATA_PATH)
assert "celltype_major" in xenium_sdata["table"].obs, (
"The Xenium data does not contain the cell types annotation; it seems that it refers to the Xenium "
"Zarr data that has not been processed with the analysis notebook mentioned in the 'Getting the Zarr"
"files' section."
)
Let’s create a new SpatialData object with just the elements we are interest in. We will predict the Xenium cell types from the Visium image, so let’s grab the cell circles and the table from the Xenium data, and the full resolution H&E image from Visium.
merged = SpatialData(
images={
"CytAssist_FFPE_Human_Breast_Cancer_full_image": visium_sdata.images[
"CytAssist_FFPE_Human_Breast_Cancer_full_image"
],
},
shapes={
"cell_circles": xenium_sdata.shapes["cell_circles"],
"cell_boundaries": xenium_sdata.shapes["cell_boundaries"],
},
tables={"table": xenium_sdata["table"]},
)
For the sake of reducing the computational requirements to run this example, let’s spatially subset the data.
min_coordinate = [12790, 12194]
max_coordinate = [15100, 15221]
merged = merged.query.bounding_box(
min_coordinate=min_coordinate,
max_coordinate=max_coordinate,
axes=["y", "x"],
target_coordinate_system="aligned",
)
visium_sdata
SpatialData object with:
├── Images
│ ├── 'CytAssist_FFPE_Human_Breast_Cancer_full_image': MultiscaleSpatialImage[cyx] (3, 21571, 19505), (3, 10785, 9752), (3, 5392, 4876), (3, 2696, 2438), (3, 1348, 1219)
│ ├── 'CytAssist_FFPE_Human_Breast_Cancer_hires_image': SpatialImage[cyx] (3, 2000, 1809)
│ └── 'CytAssist_FFPE_Human_Breast_Cancer_lowres_image': SpatialImage[cyx] (3, 600, 543)
├── Shapes
│ ├── 'CytAssist_FFPE_Human_Breast_Cancer': GeoDataFrame shape: (4992, 2) (2D shapes)
│ └── 'visium_landmarks': GeoDataFrame shape: (3, 2) (2D shapes)
└── Tables
└── 'table': AnnData (4992, 18085)
with coordinate systems:
▸ 'aligned', with elements:
CytAssist_FFPE_Human_Breast_Cancer_full_image (Images), CytAssist_FFPE_Human_Breast_Cancer (Shapes), visium_landmarks (Shapes)
▸ 'downscaled_hires', with elements:
CytAssist_FFPE_Human_Breast_Cancer_hires_image (Images), CytAssist_FFPE_Human_Breast_Cancer (Shapes)
▸ 'downscaled_lowres', with elements:
CytAssist_FFPE_Human_Breast_Cancer_lowres_image (Images), CytAssist_FFPE_Human_Breast_Cancer (Shapes)
▸ 'global', with elements:
CytAssist_FFPE_Human_Breast_Cancer_full_image (Images), CytAssist_FFPE_Human_Breast_Cancer (Shapes), visium_landmarks (Shapes)
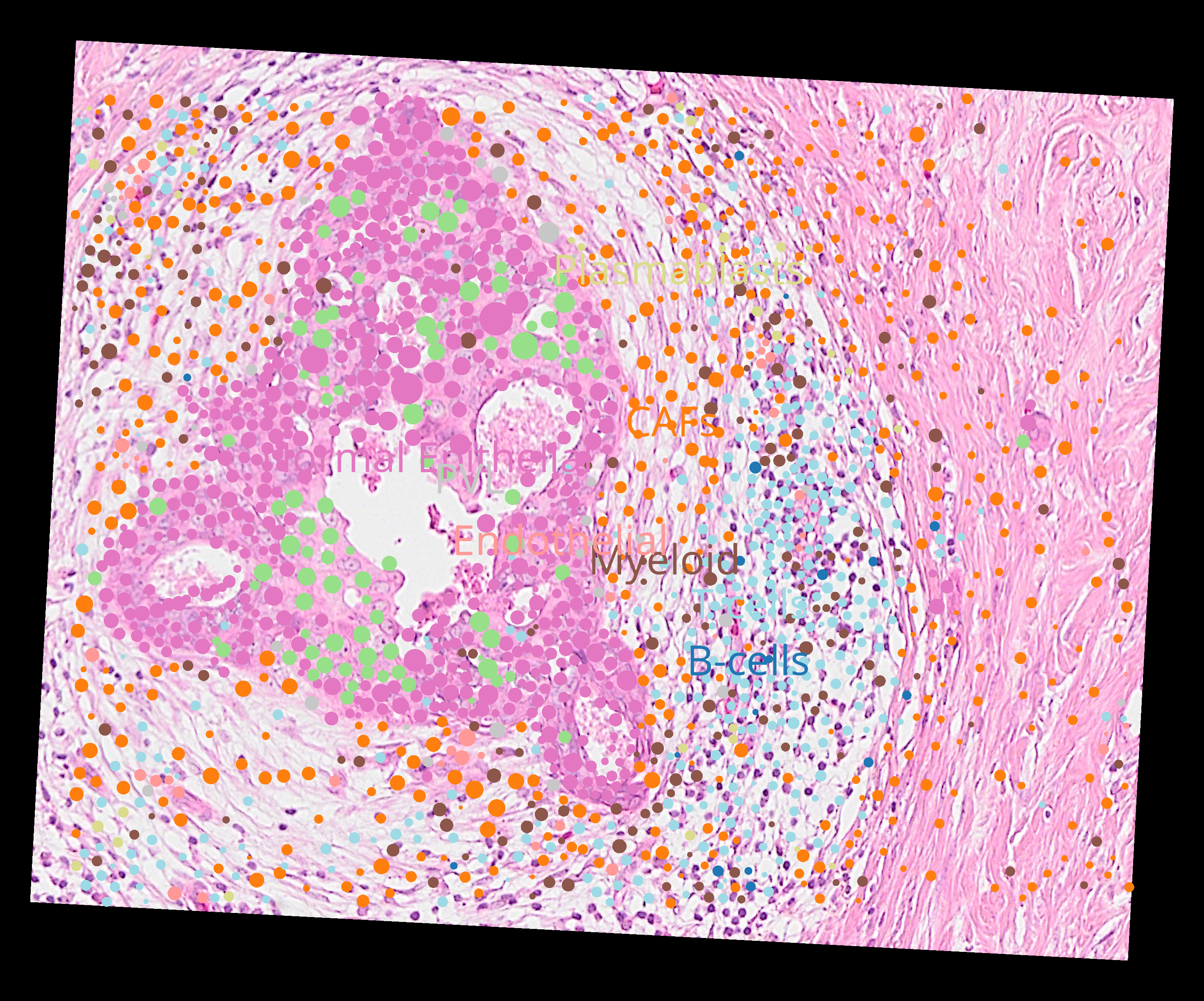
Here is a visualization of the image and cell type data. Notice how the Visium image is rotated with respect to the Xenium data.
Let’s compute the mean Xenium cell diamater, we will use this to choose an appropriate image tile size.
circles = merged["cell_circles"]
transformed_circles = transform(circles, to_coordinate_system="aligned")
xenium_circles_diameter = 2 * np.mean(transformed_circles.radius)
Let’s find the list of all the cell types we are dealing with.
cell_types = merged["table"].obs["celltype_major"].cat.categories.tolist()
We now effortlessly define a PyTorch Dataset for the SpatialData object using the class ImageTileDataset().
In particular we want the following.
We want the tile size to be 32 x 32 pixels.
At the same time, we want each tile to have a spatial extent of 3 times the average Xenium cell diameter
For each tile we want to extract the value of the
celltype_majorcategorical column and encode this into a one-hot vector. We will use thetorchvisiontransforms paradigma for achieving this.
Technical note.
There are some limitations when using PyTorch inside a Jupyter Notebook. Here we would need a function, that we call my_transform(), that we would use to apply a data transformation to the dataset. The function can’t be defined here in the notebook so we will import it from a separate Python file. For more details please see here: https://stackoverflow.com/a/65001152.
Here is the function that we would like to define.
def my_transform(sdata: SpatialData) -> tuple[torch.tensor, torch.tensor]:
tile = sdata['CytAssist_FFPE_Human_Breast_Cancer_full_image'].data.compute()
tile = torch.tensor(tile)
expected_category = sdata["table"].obs['celltype_major'].values[0]
expected_category = cell_types.index(expected_category)
cell_type = F.one_hot(
torch.tensor(expected_category), num_classes=len(cell_types)
)
return tile, cell_type
# let's import the above function
from densenet_utils import my_transform
dataset = ImageTilesDataset(
sdata=merged,
regions_to_images={"cell_circles": "CytAssist_FFPE_Human_Breast_Cancer_full_image"},
regions_to_coordinate_systems={"cell_circles": "aligned"},
table_name="table",
tile_dim_in_units=3 * xenium_circles_diameter,
transform=my_transform,
rasterize=True,
rasterize_kwargs={"target_width": 32},
)
dataset[0]
(tensor([[[243., 255., 252., ..., 255., 255., 255.],
[252., 255., 250., ..., 253., 254., 255.],
[255., 255., 250., ..., 250., 252., 255.],
...,
[255., 255., 255., ..., 255., 251., 255.],
[249., 254., 253., ..., 255., 250., 252.],
[241., 251., 249., ..., 255., 248., 255.]],
[[170., 187., 197., ..., 201., 209., 183.],
[183., 190., 195., ..., 195., 199., 182.],
[195., 194., 201., ..., 187., 200., 177.],
...,
[198., 206., 203., ..., 218., 222., 176.],
[188., 197., 196., ..., 222., 221., 175.],
[180., 191., 192., ..., 224., 220., 181.]],
[[216., 231., 226., ..., 225., 238., 237.],
[227., 227., 224., ..., 220., 238., 235.],
[231., 225., 223., ..., 214., 238., 229.],
...,
[235., 235., 234., ..., 237., 242., 213.],
[222., 230., 229., ..., 240., 239., 211.],
[213., 225., 225., ..., 240., 235., 217.]]]),
tensor([0., 1., 0., 0., 0., 0., 0., 0., 0.]))
Let’s now define a PyTorch Lightning data module to reduce the amount of boilerplate code we need to write.
class TilesDataModule(LightningDataModule):
def __init__(self, batch_size: int, num_workers: int, dataset: torch.utils.data.Dataset):
super().__init__()
self.batch_size = batch_size
self.num_workers = num_workers
self.dataset = dataset
def setup(self, stage=None):
n_train = int(len(self.dataset) * 0.7)
n_val = int(len(self.dataset) * 0.2)
n_test = len(self.dataset) - n_train - n_val
self.train, self.val, self.test = torch.utils.data.random_split(
self.dataset,
[n_train, n_val, n_test],
generator=torch.Generator().manual_seed(42),
)
def train_dataloader(self):
return DataLoader(
self.train,
batch_size=self.batch_size,
num_workers=self.num_workers,
shuffle=True,
)
def val_dataloader(self):
return DataLoader(
self.val,
batch_size=self.batch_size,
num_workers=self.num_workers,
shuffle=False,
)
def test_dataloader(self):
return DataLoader(
self.test,
batch_size=self.batch_size,
num_workers=self.num_workers,
shuffle=False,
)
def predict_dataloader(self):
return DataLoader(
self.dataset,
batch_size=self.batch_size,
num_workers=self.num_workers,
shuffle=False,
)
Let’s define the Dense Net, that we import from Monai.
class DenseNetModel(pl.LightningModule):
def __init__(self, learning_rate: float, in_channels: int, num_classes: int):
super().__init__()
# store hyperparameters
self.save_hyperparameters()
self.loss_function = CrossEntropyLoss()
# make the model
self.model = DenseNet121(spatial_dims=2, in_channels=in_channels, out_channels=num_classes)
def forward(self, x) -> torch.Tensor:
return self.model(x)
def _compute_loss_from_batch(self, batch: Dict[str, torch.Tensor], batch_idx: int) -> float:
inputs = batch[0]
labels = batch[1]
outputs = self.model(inputs)
return self.loss_function(outputs, labels)
def training_step(self, batch: Dict[str, torch.Tensor], batch_idx: int) -> Dict[str, float]:
# compute the loss
loss = self._compute_loss_from_batch(batch=batch, batch_idx=batch_idx)
# perform logging
self.log("training_loss", loss, batch_size=len(batch[0]))
return {"loss": loss}
def validation_step(self, batch: Dict[str, torch.Tensor], batch_idx: int) -> float:
loss = self._compute_loss_from_batch(batch=batch, batch_idx=batch_idx)
imgs, labels = batch
acc = self.compute_accuracy(imgs, labels)
# By default logs it per epoch (weighted average over batches), and returns it afterwards
self.log("test_acc", acc)
return loss
def test_step(self, batch, batch_idx):
imgs, labels = batch
acc = self.compute_accuracy(imgs, labels)
# By default logs it per epoch (weighted average over batches), and returns it afterwards
self.log("test_acc", acc)
def predict_step(self, batch, batch_idx: int, dataloader_idx: int = 0):
imgs, labels = batch
preds = self.model(imgs).argmax(dim=-1)
return preds
def compute_accuracy(self, imgs, labels):
preds = self.model(imgs).argmax(dim=-1)
labels_value = torch.argmax(labels, dim=-1)
acc = (labels_value == preds).float().mean()
return acc
def configure_optimizers(self) -> Adam:
return Adam(self.model.parameters(), lr=self.hparams.learning_rate)
We are ready to train the model!
import os
import numpy as np
import pytorch_lightning as pl
import torch
import torchvision
from pytorch_lightning.callbacks import LearningRateMonitor
from pytorch_lightning.callbacks.progress import TQDMProgressBar
from pytorch_lightning.loggers import CSVLogger
pl.seed_everything(7)
PATH_DATASETS = os.environ.get("PATH_DATASETS", "..")
BATCH_SIZE = 4096 if torch.cuda.is_available() else 64
NUM_WORKERS = 10 if torch.cuda.is_available() else 8
print(f"Using {BATCH_SIZE} batch size.")
print(f"Using {NUM_WORKERS} workers.")
tiles_data_module = TilesDataModule(batch_size=BATCH_SIZE, num_workers=NUM_WORKERS, dataset=dataset)
tiles_data_module.setup()
train_dl = tiles_data_module.train_dataloader()
val_dl = tiles_data_module.val_dataloader()
test_dl = tiles_data_module.test_dataloader()
num_classes = len(cell_types)
in_channels = dataset[0][0].shape[0]
model = DenseNetModel(
learning_rate=1e-5,
in_channels=in_channels,
num_classes=num_classes,
)
import logging
logging.basicConfig(level=logging.INFO)
trainer = pl.Trainer(
max_epochs=2,
accelerator="auto",
# devices=1, # limiting got iPython runs. Edit: it works also without now
logger=CSVLogger(save_dir="logs/"),
callbacks=[
LearningRateMonitor(logging_interval="step"),
TQDMProgressBar(refresh_rate=5),
],
log_every_n_steps=20,
)
Using 64 batch size.
Using 8 workers.
trainer.fit(model, datamodule=tiles_data_module)
trainer.test(model, datamodule=tiles_data_module)
Epoch 0: 100%|████████████████████████████████████████████████████████████████████████████████| 20/20 [00:28<00:00, 0.71it/s, v_num=34]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: 0%| | 0/6 [00:00<?, ?it/s]
Validation DataLoader 0: 0%| | 0/6 [00:00<?, ?it/s]
Validation DataLoader 0: 83%|███████████████████████████████████████████████████████████████▎ | 5/6 [00:01<00:00, 2.76it/s]
Validation DataLoader 0: 100%|████████████████████████████████████████████████████████████████████████████| 6/6 [00:02<00:00, 2.84it/s]
Epoch 1: 100%|████████████████████████████████████████████████████████████████████████████████| 20/20 [00:51<00:00, 0.39it/s, v_num=34]
Validation: | | 0/? [00:00<?, ?it/s]
Validation: 0%| | 0/6 [00:00<?, ?it/s]
Validation DataLoader 0: 0%| | 0/6 [00:00<?, ?it/s]
Validation DataLoader 0: 83%|███████████████████████████████████████████████████████████████▎ | 5/6 [00:01<00:00, 3.07it/s]
Validation DataLoader 0: 100%|████████████████████████████████████████████████████████████████████████████| 6/6 [00:01<00:00, 3.42it/s]
Epoch 1: 100%|████████████████████████████████████████████████████████████████████████████████| 20/20 [01:19<00:00, 0.25it/s, v_num=34]Epoch 1: 100%|████████████████████████████████████████████████████████████████████████████████| 20/20 [01:19<00:00, 0.25it/s, v_num=34]
Testing DataLoader 0: 100%|███████████████████████████████████████████████████████████████████████████████| 3/3 [00:00<00:00, 8.50it/s]
┏━━━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━━━━━━━┓ ┃ Test metric ┃ DataLoader 0 ┃ ┡━━━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━━━━━━━┩ │ test_acc │ 0.3186813294887543 │ └───────────────────────────┴───────────────────────────┘
[{'test_acc': 0.3186813294887543}]
# model = DenseNetModel.load_from_checkpoint('logs/lightning_logs/version_12/checkpoints/epoch=1-step=40.ckpt')
# disable randomness, dropout, etc...
model.eval()
trainer = pl.Trainer(
accelerator="auto",
devices=1,
callbacks=[
TQDMProgressBar(refresh_rate=10),
],
)
predictions = trainer.predict(datamodule=tiles_data_module, model=model)
predictions = torch.cat(predictions, dim=0)
print(np.unique(predictions.detach().cpu().numpy(), return_counts=True))
Predicting DataLoader 0: 100%|██████████████████████████████████████████████████████████████████████████| 29/29 [00:34<00:00, 0.84it/s]
(array([0, 1, 2, 3, 4, 5, 6, 7, 8]), array([ 66, 430, 65, 88, 313, 246, 21, 20, 563]))
p = predictions.detach().cpu().numpy()
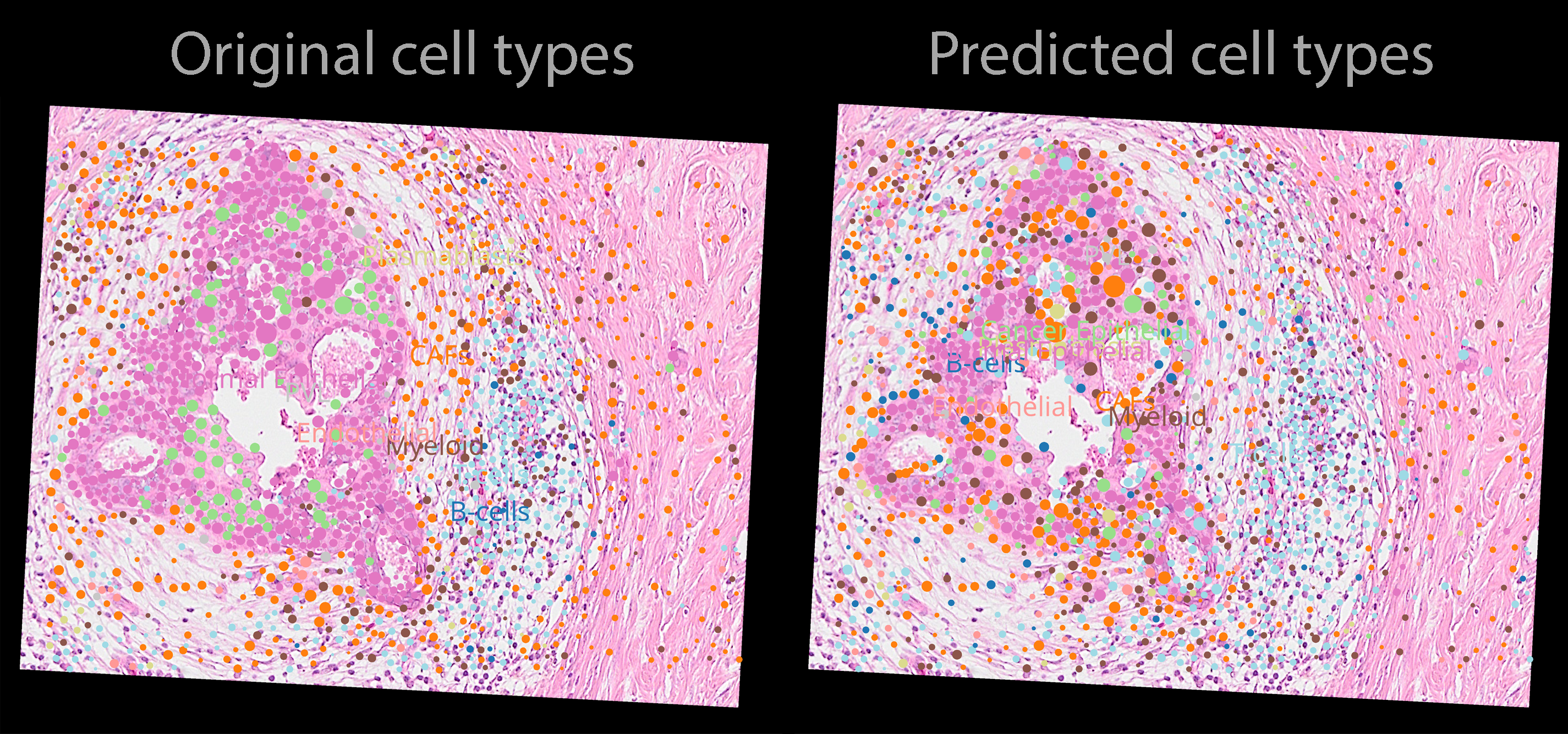
predicted_celltype_major = []
for i in p:
predicted_celltype_major.append(cell_types[i])
s = pd.Series(predicted_celltype_major)
categorical = pd.Categorical(s, categories=cell_types)
categorical.index = merged["table"].obs.index
merged["table"].obs["predicted_celltype_major"] = categorical
Here are the precitions from the model (napari screenshot).
merged
SpatialData object with:
├── Images
│ └── 'CytAssist_FFPE_Human_Breast_Cancer_full_image': MultiscaleSpatialImage[cyx] (3, 1213, 952), (3, 607, 476), (3, 303, 238), (3, 152, 119), (3, 76, 60)
├── Shapes
│ ├── 'cell_boundaries': GeoDataFrame shape: (1899, 1) (2D shapes)
│ └── 'cell_circles': GeoDataFrame shape: (1812, 2) (2D shapes)
└── Tables
└── 'table': AnnData (1812, 313)
with coordinate systems:
▸ 'aligned', with elements:
CytAssist_FFPE_Human_Breast_Cancer_full_image (Images), cell_boundaries (Shapes), cell_circles (Shapes)
▸ 'global', with elements:
CytAssist_FFPE_Human_Breast_Cancer_full_image (Images), cell_boundaries (Shapes), cell_circles (Shapes)
adata_polygons = merged["table"].copy()
adata_polygons.uns["spatialdata_attrs"]["region"] = "cell_boundaries"
adata_polygons.obs["region"] = "cell_boundaries"
adata_polygons.obs["region"] = adata_polygons.obs["region"].astype("category")
del merged.tables["table"]
merged["table"] = adata_polygons
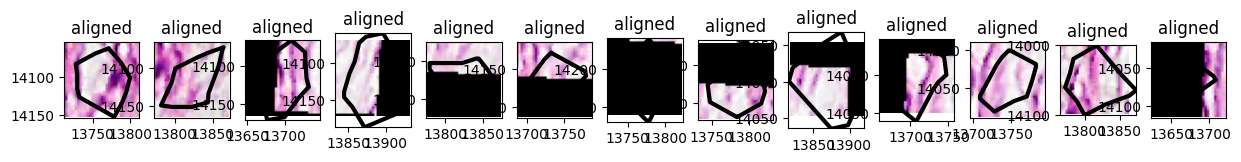
Visualizing the tiles#
x = np.array([13694.0, 13889.0, 13889.0, 13694.0, 13694.0])
y = np.array([13984.0, 13984.0, 14162.0, 14162.0, 13984.0])
small_sdata = merged.query.bounding_box(
axes=("x", "y"),
min_coordinate=[np.min(x), np.min(y)],
max_coordinate=[np.max(x), np.max(y)],
target_coordinate_system="aligned",
)
small_sdata
SpatialData object with:
├── Images
│ └── 'CytAssist_FFPE_Human_Breast_Cancer_full_image': MultiscaleSpatialImage[cyx] (3, 79, 73), (3, 40, 36), (3, 20, 18), (3, 10, 9), (3, 5, 4)
├── Shapes
│ ├── 'cell_boundaries': GeoDataFrame shape: (13, 1) (2D shapes)
│ └── 'cell_circles': GeoDataFrame shape: (8, 2) (2D shapes)
└── Tables
└── 'table': AnnData (13, 313)
with coordinate systems:
▸ 'aligned', with elements:
CytAssist_FFPE_Human_Breast_Cancer_full_image (Images), cell_boundaries (Shapes), cell_circles (Shapes)
▸ 'global', with elements:
CytAssist_FFPE_Human_Breast_Cancer_full_image (Images), cell_boundaries (Shapes), cell_circles (Shapes)
small_dataset = ImageTilesDataset(
sdata=small_sdata,
regions_to_images={"cell_boundaries": "CytAssist_FFPE_Human_Breast_Cancer_full_image"},
regions_to_coordinate_systems={"cell_boundaries": "aligned"},
tile_dim_in_units=100,
rasterize=True,
rasterize_kwargs={"target_width": 32},
table_name="table",
transform=None,
)
small_dataset[0]
SpatialData object with:
├── Images
│ └── 'CytAssist_FFPE_Human_Breast_Cancer_full_image': SpatialImage[cyx] (3, 32, 32)
└── Tables
└── 'table': AnnData (1, 313)
with coordinate systems:
▸ 'aligned', with elements:
CytAssist_FFPE_Human_Breast_Cancer_full_image (Images)
import matplotlib.pyplot as plt
import spatialdata as sd
import spatialdata_plot
from geopandas import GeoDataFrame
from spatialdata.models import ShapesModel
n = len(small_dataset)
axes = plt.subplots(1, n, figsize=(15, 3))[1]
for sdata_tile, i in zip(small_dataset, range(n)):
region, instance_id = small_dataset.dataset_index.iloc[i][["region", "instance_id"]]
shapes = small_sdata[region]
transformations = get_transformation(shapes, get_all=True)
tile = ShapesModel.parse(GeoDataFrame(geometry=shapes.loc[instance_id]), transformations=transformations)
# BUG: we need to explicitly remove the coordinate system global if we want to combine
# images and shapes plots into a single subplot
# https://github.com/scverse/spatialdata-plot/issues/176
sdata_tile["cell_boundaries"] = tile
if "global" in get_transformation(sdata_tile["cell_boundaries"], get_all=True):
sd.transformations.remove_transformation(sdata_tile["cell_boundaries"], "global")
sdata_tile.pl.render_images().pl.render_shapes(
# outline_color='predicted_celltype_major', # not yet supported: https://github.com/scverse/spatialdata-plot/issues/137
outline_width=3.0,
outline=True,
fill_alpha=0.0,
).pl.show(
ax=axes[i],
)