Annotating regions of interest with napari#
We will show:
how to use
naparifor drawing regions of interesets and how to access these annotations via code.assuming these saved regions are anatomical regions of interterest, how to extract the Visium spots/cells under these regions.
# %load_ext jupyter_black
import numpy as np
import pandas as pd
import spatialdata as sd
from geopandas import GeoDataFrame
from shapely import Polygon
from spatialdata.models import ShapesModel
from spatialdata.transformations import Identity
# from napari_spatialdata import Interactive
You can download the processed Visium data (already aligned) from here: Visium dataset. In alternative you can obtain the data by running this analysis notebook.
Please rename the file to visium_aligned.zarr and place it in the same folder as this notebook (or use symlinks to make the data accessible).
visium_sdata = sd.read_zarr("visium_aligned.zarr")
Adding shapes annotations#
We can use napari to draw rectangles and generic polygons.
This is the procedure to annotate and save the shapes (shown in the GIF):
open
napariwithInteractive()fromnapari_spatialdatacreate a new Shapes layer in napari
(optional) rename the layer
(optional) change the fill and border propoerties for easier visualization
select the “Rectangle tool” or the “Polygon tool”
(optional) use the
naparifunctions to move/delete/modify the shapes (shown later)save the annotation to the
SpatialDataobject by pressingShift + E(if you calledInteractive()passing multipleSpatialDataobjects, the annotations will be saved to one of them).
When pressing Shift + E, the shapes are saved. Anyway, for making this notebook reproducible, let’s manually hardcode the shapes that have been drawn in the GIF above. The code produces data that is equivalent to the one given by napari. Note that we could have also put the shapes in a new SpatialData objects, saved this to Zarr and loaded it here.
shape0 = np.array([[7758.0, 11577.0, 11577.0, 7758.0, 7758.0], [10928.0, 10928.0, 17197.0, 17197.0, 10928.0]])
shape1 = np.array([[11721.0, 14712.0, 14712.0, 11721.0, 11721.0], [12117.0, 12117.0, 16801.0, 16801.0, 12117.0]])
# fmt: off
shape2 = np.array(
[
[
31320.0,28114.0,26745.0,26060.0,22998.0,23863.0,23899.0,29159.0,29195.0,32113.0,31789.0,33230.0,31320.0,
],
[
23286.0,20764.0,20836.0,19215.0,18170.0,15756.0,12081.0,12766.0,14567.0,17125.0,18530.0,20692.0,23286.0,
],
]
)
# fmt: on
def numpy_to_shapely(x: np.array) -> Polygon:
return Polygon(list(map(tuple, x.T)))
gdf = GeoDataFrame({"geometry": [numpy_to_shapely(shape0), numpy_to_shapely(shape1), numpy_to_shapely(shape2)]})
gdf = ShapesModel.parse(gdf, transformations={"aligned": Identity()})
visium_sdata.shapes["my_shapes"] = gdf
Importantly, notice how the shapes "my_shapes", now available in the SpatialData object, are aligned to the "aligned" coordinate system. This is because the shapes have been drawn when the coordinate system "aligned" was selected in napari.
visium_sdata
SpatialData object with:
├── Images
│ ├── 'CytAssist_FFPE_Human_Breast_Cancer_full_image': MultiscaleSpatialImage[cyx] (3, 21571, 19505), (3, 10785, 9752), (3, 5392, 4876), (3, 2696, 2438), (3, 1348, 1219)
│ ├── 'CytAssist_FFPE_Human_Breast_Cancer_hires_image': SpatialImage[cyx] (3, 2000, 1809)
│ └── 'CytAssist_FFPE_Human_Breast_Cancer_lowres_image': SpatialImage[cyx] (3, 600, 543)
├── Shapes
│ ├── 'CytAssist_FFPE_Human_Breast_Cancer': GeoDataFrame shape: (4992, 2) (2D shapes)
│ └── 'my_shapes': GeoDataFrame shape: (3, 1) (2D shapes)
└── Tables
└── 'table': AnnData (4992, 18085)
with coordinate systems:
▸ 'aligned', with elements:
CytAssist_FFPE_Human_Breast_Cancer_full_image (Images), CytAssist_FFPE_Human_Breast_Cancer (Shapes), my_shapes (Shapes)
▸ 'downscaled_hires', with elements:
CytAssist_FFPE_Human_Breast_Cancer_hires_image (Images), CytAssist_FFPE_Human_Breast_Cancer (Shapes)
▸ 'downscaled_lowres', with elements:
CytAssist_FFPE_Human_Breast_Cancer_lowres_image (Images), CytAssist_FFPE_Human_Breast_Cancer (Shapes)
▸ 'global', with elements:
CytAssist_FFPE_Human_Breast_Cancer_full_image (Images), CytAssist_FFPE_Human_Breast_Cancer (Shapes)
The shapes are stored in a geopandas.GeoDataFrame as shapely.Polygon objects. Hence we can use them in any geopandas/shapely workflow.
visium_sdata["my_shapes"]
| geometry | |
|---|---|
| 0 | POLYGON ((7758.000 10928.000, 11577.000 10928.... |
| 1 | POLYGON ((11721.000 12117.000, 14712.000 12117... |
| 2 | POLYGON ((31320.000 23286.000, 28114.000 20764... |
We can quickly see the shapes in the notebook (thanks to shapely). For more complex visualization involving different elements or different coordinate system we can rely on spatialdata-plot.
visium_sdata["my_shapes"].geometry.iloc[0]
visium_sdata["my_shapes"].geometry.iloc[2]
Manipulating the shapes in napari#
Here is an example of how we can use napari to move/modify/delete some of the shapes, and save them to another layer (in-place overwrite is also supported).
Interactive(visium_sdata)
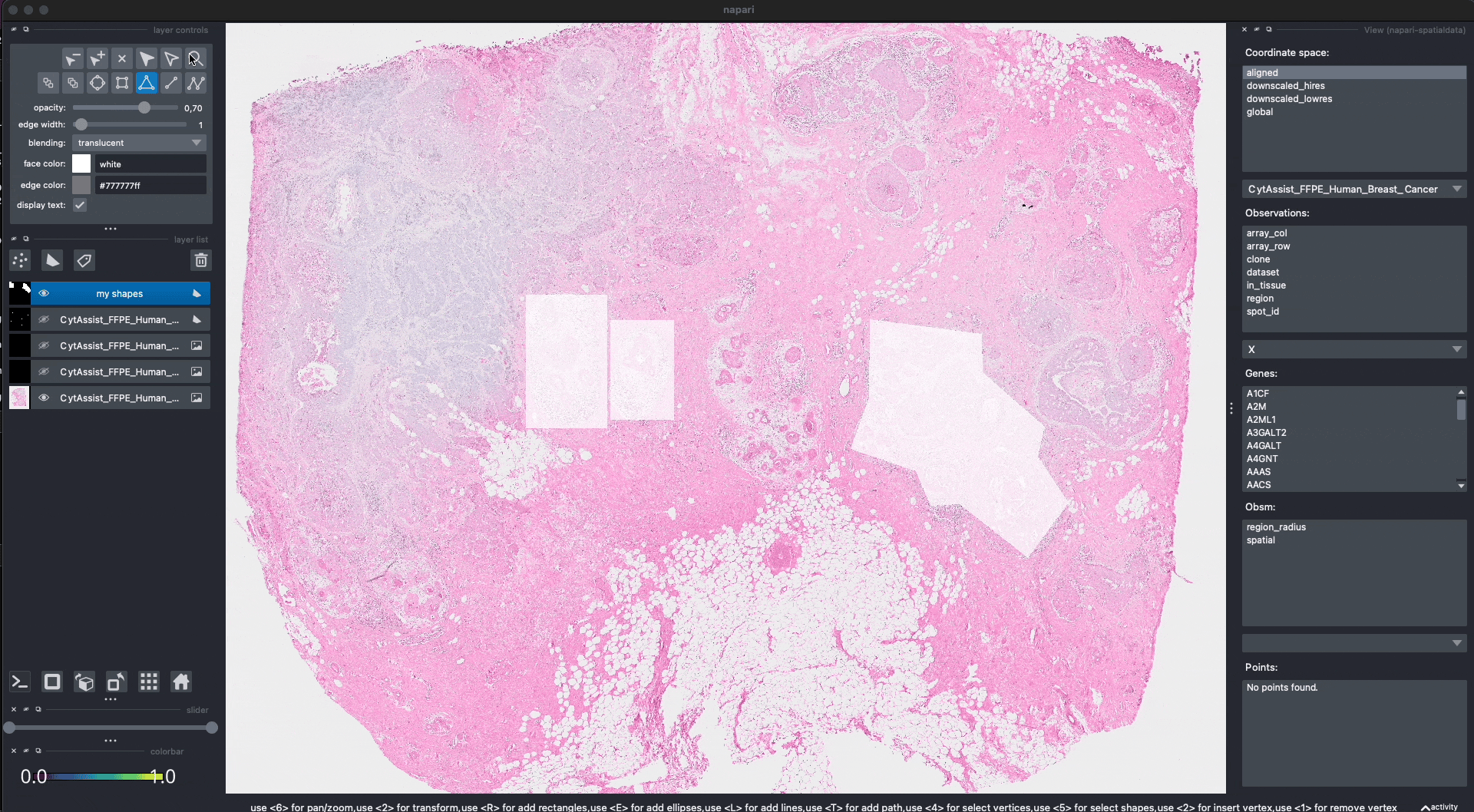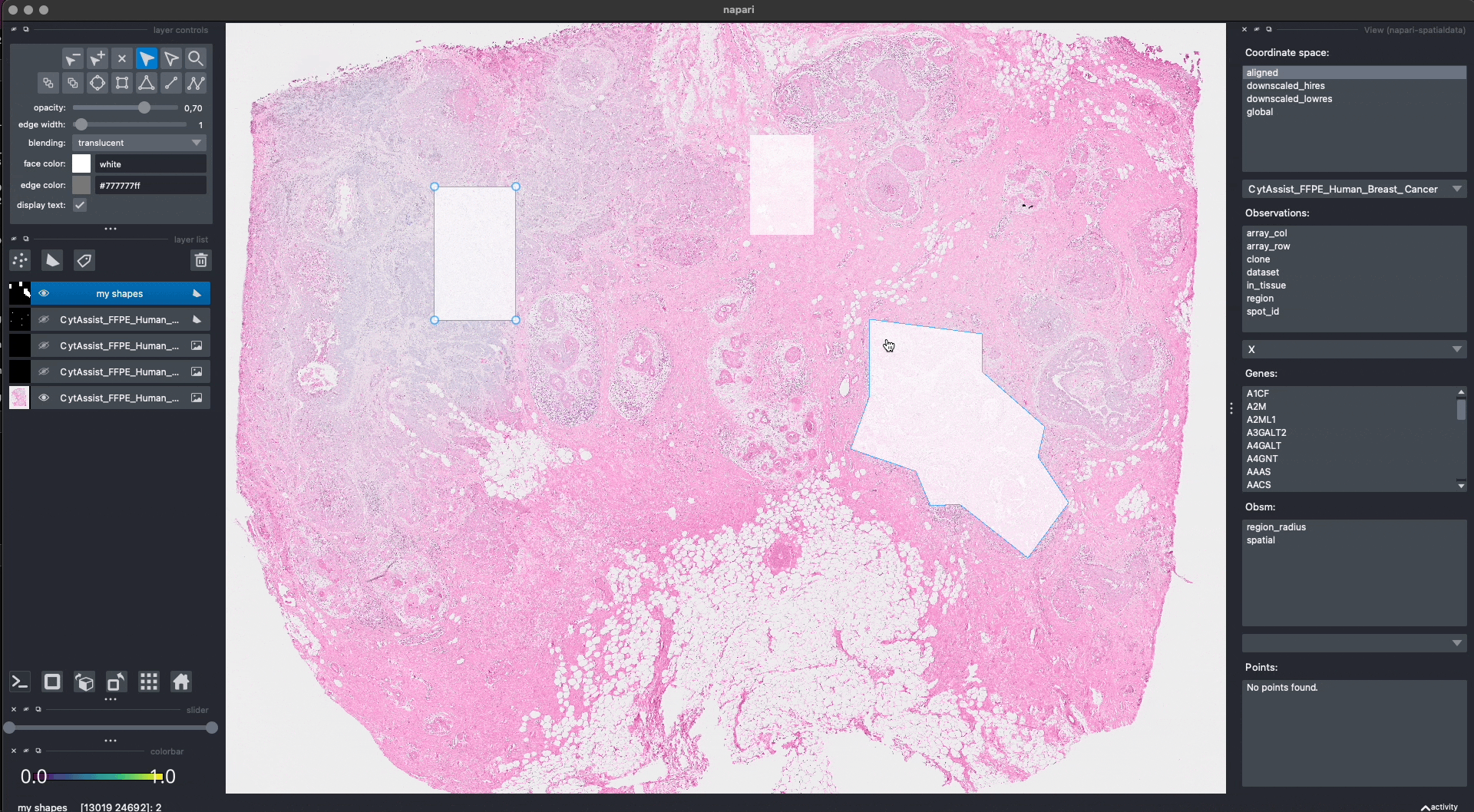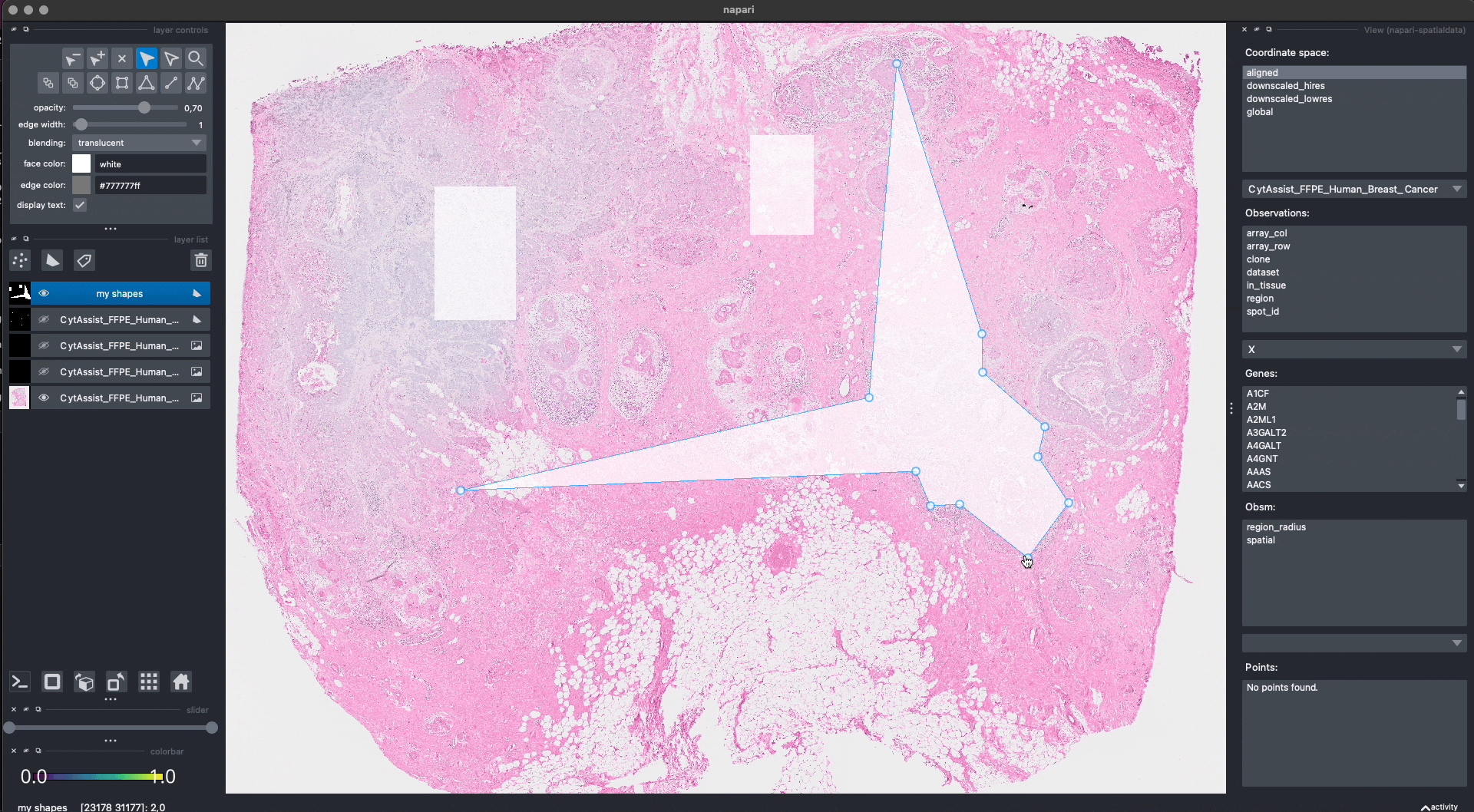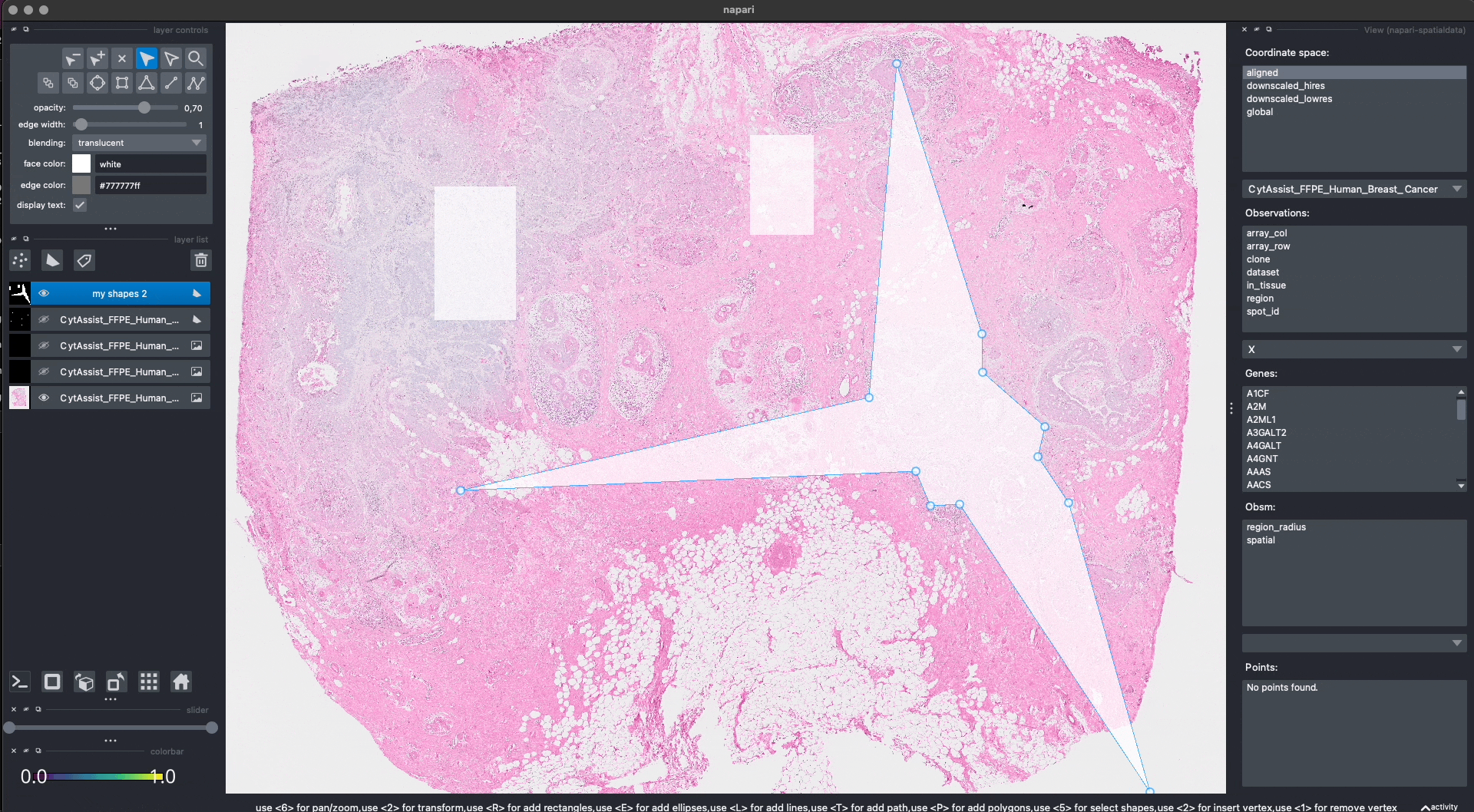
We saved the new layer as my_shapes_2 inside the same SpatialData object (again using the "aligned" coordinate system).
As above, let’s hardcode the shapes to make this notebook reproducible.
shape0 = np.array([[3471.0, 7290.0, 7290.0, 3471.0, 3471.0], [5848.0, 5848.0, 12117.0, 12117.0, 5848.0]])
shape1 = np.array([[18278.0, 21269.0, 21269.0, 18278.0, 18278.0], [3435.0, 3435.0, 8118.0, 8118.0, 3435.0]])
# fmt: off
shape2 = np.array(
[
[
37049.0,28114.0,26745.0,26060.0,4696.0,23863.0,25160.0,29159.0,29195.0,32113.0,31789.0,33230.0,37049.0,
],
[
34274.0,20764.0,20836.0,19215.0,20116.0,15756.0,84.0,12766.0,14567.0,17125.0,18530.0,20692.0,34274.0,
],
]
)
# fmt: on
gdf = GeoDataFrame({"geometry": [numpy_to_shapely(shape0), numpy_to_shapely(shape1), numpy_to_shapely(shape2)]})
gdf = ShapesModel.parse(gdf, transformations={"aligned": Identity()})
visium_sdata.shapes["my_shapes_2"] = gdf
visium_sdata
SpatialData object with:
├── Images
│ ├── 'CytAssist_FFPE_Human_Breast_Cancer_full_image': MultiscaleSpatialImage[cyx] (3, 21571, 19505), (3, 10785, 9752), (3, 5392, 4876), (3, 2696, 2438), (3, 1348, 1219)
│ ├── 'CytAssist_FFPE_Human_Breast_Cancer_hires_image': SpatialImage[cyx] (3, 2000, 1809)
│ └── 'CytAssist_FFPE_Human_Breast_Cancer_lowres_image': SpatialImage[cyx] (3, 600, 543)
├── Shapes
│ ├── 'CytAssist_FFPE_Human_Breast_Cancer': GeoDataFrame shape: (4992, 2) (2D shapes)
│ ├── 'my_shapes': GeoDataFrame shape: (3, 1) (2D shapes)
│ └── 'my_shapes_2': GeoDataFrame shape: (3, 1) (2D shapes)
└── Tables
└── 'table': AnnData (4992, 18085)
with coordinate systems:
▸ 'aligned', with elements:
CytAssist_FFPE_Human_Breast_Cancer_full_image (Images), CytAssist_FFPE_Human_Breast_Cancer (Shapes), my_shapes (Shapes), my_shapes_2 (Shapes)
▸ 'downscaled_hires', with elements:
CytAssist_FFPE_Human_Breast_Cancer_hires_image (Images), CytAssist_FFPE_Human_Breast_Cancer (Shapes)
▸ 'downscaled_lowres', with elements:
CytAssist_FFPE_Human_Breast_Cancer_lowres_image (Images), CytAssist_FFPE_Human_Breast_Cancer (Shapes)
▸ 'global', with elements:
CytAssist_FFPE_Human_Breast_Cancer_full_image (Images), CytAssist_FFPE_Human_Breast_Cancer (Shapes)
visium_sdata["my_shapes_2"].geometry.iloc[2]
Lasso annotations#
We are working with the napari developers for enabling a lasso tool for drawing anatomical annotations. This functionality will be part of napari as a new button in the user interface. With SpatialData it will be possible to save and represent the annotatoins in-memory, as shown above. The lasso tool will have support for both mouse/trackpad and graphic tables.
In the GIF you can see a preview, the experimental code is available at https://github.com/napari/napari/pull/5555.
# we rounded the coordinates to make it less verbose here in the notebook
# fmt: off
shape0 = (
np.array(
[
[
182.0,181.0,179.0,178.0,174.0,173.0,172.0,168.0,165.0,164.0,164.0,166.0,167.0,168.0,169.0,169.0,168.0,166.0,163.0,163.0,
163.0,164.0,165.0,167.0,168.0,169.0,172.0,173.0,174.0,175.0,180.0,185.0,187.0,188.0,189.0,190.0,191.0,192.0,193.0,194.0,
196.0,198.0,200.0,202.0,205.0,207.0,209.0,211.0,212.0,213.0,215.0,215.0,216.0,216.0,215.0,215.0,214.0,212.0,211.0,209.0,
208.0,208.0,206.0,205.0,204.0,202.0,200.0,196.0,195.0,195.0,195.0,194.0,192.0,190.0,189.0,186.0,184.0,184.0,183.0,184.0,
185.0,186.0,186.0,184.0,182.0,182.0,182.0,183.0,183.0,183.0,182.0,182.0,183.0,183.0,181.0,178.0,178.0,178.0,179.0,180.0,
182.0,182.0,183.0,183.0,182.0,
],
[
105.0,105.0,106.0,107.0,108.0,109.0,110.0,112.0,114.0,115.0,118.0,121.0,122.0,123.0,125.0,126.0,129.0,136.0,140.0,141.0,
143.0,151.0,153.0,155.0,157.0,158.0,160.0,162.0,162.0,163.0,168.0,173.0,174.0,174.0,175.0,176.0,178.0,179.0,179.0,181.0,
181.0,182.0,183.0,183.0,183.0,183.0,182.0,181.0,179.0,177.0,174.0,173.0,170.0,167.0,164.0,160.0,159.0,158.0,159.0,161.0,
161.0,161.0,158.0,156.0,156.0,156.0,157.0,160.0,161.0,162.0,164.0,164.0,164.0,162.0,161.0,160.0,159.0,157.0,156.0,153.0,
151.0,148.0,147.0,145.0,144.0,143.0,141.0,139.0,137.0,136.0,135.0,134.0,132.0,131.0,129.0,126.0,123.0,120.0,118.0,115.0,
112.0,111.0,110.0,109.0,105.0,
],
]
)
* 100
)
# fmt: on
gdf = GeoDataFrame({"geometry": [numpy_to_shapely(shape0)]})
gdf = ShapesModel.parse(gdf, transformations={"aligned": Identity()})
visium_sdata.shapes["lasso"] = gdf
visium_sdata.shapes["lasso"].geometry.iloc[0]
Extracting cells under the regions#
Let’s now find the cells under each of the three regions of interest and record, in the table as a categorical column, which category each cell belongs.
We will use the spatial query API polygon_query(); a separate tutorial is available for it and we will further assume the reader being familiar which such functionality.
Let’s find, for each saved shape, the table of cells it contains.
from spatialdata import polygon_query
filtered_tables = {}
for shape in ["lasso", "my_shapes", "my_shapes_2"]:
polygon = visium_sdata[shape].geometry.iloc[0]
filtered_tables[shape] = polygon_query(visium_sdata, polygon=polygon, target_coordinate_system="aligned")["table"]
filtered_tables
{'lasso': AnnData object with n_obs × n_vars = 102 × 18085
obs: 'in_tissue', 'array_row', 'array_col', 'spot_id', 'region', 'dataset', 'clone'
var: 'gene_ids', 'feature_types', 'genome'
uns: 'spatial', 'spatialdata_attrs'
obsm: 'spatial',
'my_shapes': AnnData object with n_obs × n_vars = 139 × 18085
obs: 'in_tissue', 'array_row', 'array_col', 'spot_id', 'region', 'dataset', 'clone'
var: 'gene_ids', 'feature_types', 'genome'
uns: 'spatial', 'spatialdata_attrs'
obsm: 'spatial',
'my_shapes_2': AnnData object with n_obs × n_vars = 142 × 18085
obs: 'in_tissue', 'array_row', 'array_col', 'spot_id', 'region', 'dataset', 'clone'
var: 'gene_ids', 'feature_types', 'genome'
uns: 'spatial', 'spatialdata_attrs'
obsm: 'spatial'}
Note that polygon_query only returns a table in case of the table annotating an element in the
resulting queried SpatialData object. Other tables are filtered out. Should you want to keep
tables that are not annotating any elements in the resulting SpatialData object, a parameter
filter_tables set to False can be passed on to the polygon_query function. Which region(s) or
elements a table annotates can be retrieved as follows:
from spatialdata import SpatialData
# Note that we could also do visium_sdata.get_annotated regions here. This is just meant to show we
# can also get annotated regions from a table outside a Spatialdata object.
print(SpatialData.get_annotated_regions(visium_sdata["table"]))
CytAssist_FFPE_Human_Breast_Cancer
It can be that a table is annotating an element but is still not returned after the polygon query. The reason in that case is that the resulting table didn’t contain any rows annotating elements in the result of the query.
Now, let’s add a categorical column to the original full table, named 'annotation', with default categorical value 'unassigned'. Then, let’s assign, for each row in the 3 subtables, its value to the name of the relative region of interest.
# it's important for the index to be unique
assert visium_sdata["table"].obs.index.is_unique
categories = ["unassigned"] + list(filtered_tables.keys())
n = len(visium_sdata["table"])
visium_sdata["table"].obs["annotation"] = pd.Categorical(["unassigned" for _ in range(n)], categories=categories)
for shape, subtable in filtered_tables.items():
in_shape = subtable.obs.index
visium_sdata["table"].obs["annotation"].loc[in_shape] = shape
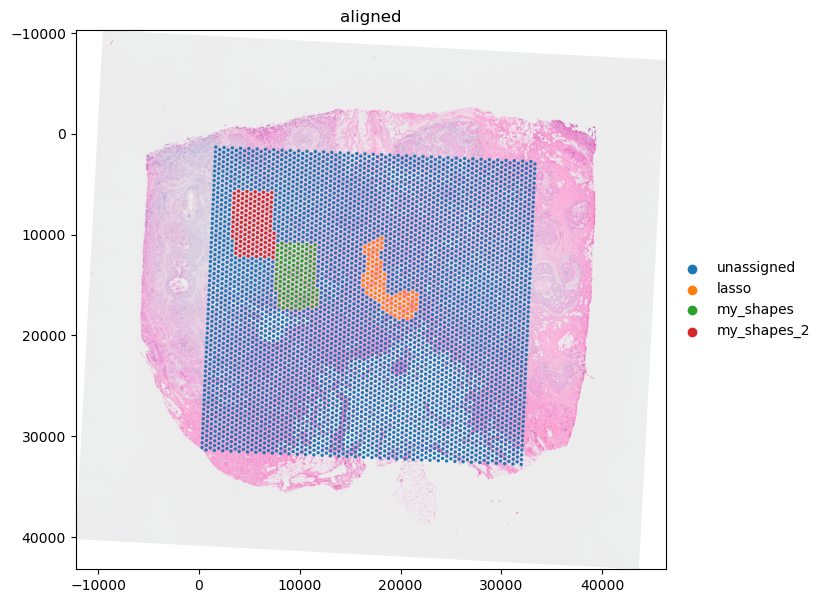
visium_sdata["table"].obs["annotation"].value_counts()
annotation
unassigned 4609
my_shapes_2 142
my_shapes 139
lasso 102
Name: count, dtype: int64
Let’s visualize the result.
import matplotlib.pyplot as plt
import spatialdata_plot
plt.figure(figsize=(12, 7))
ax = plt.gca()
visium_sdata.pl.render_images("CytAssist_FFPE_Human_Breast_Cancer_full_image").pl.render_shapes(
"CytAssist_FFPE_Human_Breast_Cancer", color="annotation"
).pl.show(coordinate_systems="aligned", ax=ax)
Limitations#
Currently we support saving rectangles, polygons and points with napari (see the landmark tutorial). We currently don’t support saving circles, ellipses or segments as well as overwriting existing elements. The latter will be possible very soon though. Furthermore, we ignore the style parameters that napari provides, such us fill color, border color, points size and line width. If you are interested in saving these types of annotations please get in touch with us or join the NGFF discussions on annotations in GitHub.
